Open access Analysis of the canid Y-chromosome phylogeny using short-read sequencing data reveals the presence of distinct haplogroups among Neolithic European dogs, by Oetjens et al., BMC Genomics (2018) 19:350.
Interesting excerpts (modified for clarity, emphasis mine):
Introduction
Canid mitochondrial phylogenies show that dogs and wolves are not reciprocally monophyletic. The mitochondrial tree contains four deeply rooted clades encompassing dogs and many grey wolf groups. These four clades form the basis of dog mitochondrial haplogroup assignment, known as haplogroups A-D. The time of the most recent common ancestor (TMRCA) of haplogroups A-D significantly predates estimates for domestication based on archeological and genetic evidence. Instead, these clades may represent variation present among the founding population of the dog lineage or the results of wolf introgressions into dog populations. The relative frequencies of mitochondria haplogroups are not stable over time, with changes reflecting processes such as drift, migration, and population growth. Although the mitochondria A and B haplogroups are most common in contemporary European dogs, surveys of ancient samples indicate that the majority of ancient European dogs carried the C or D mitochondrial haplotype. This apparent turnover in mitochondrial haplogroups may reflect the migration of a distinct dog population into Europe over the past 15,000 years.
Discussion
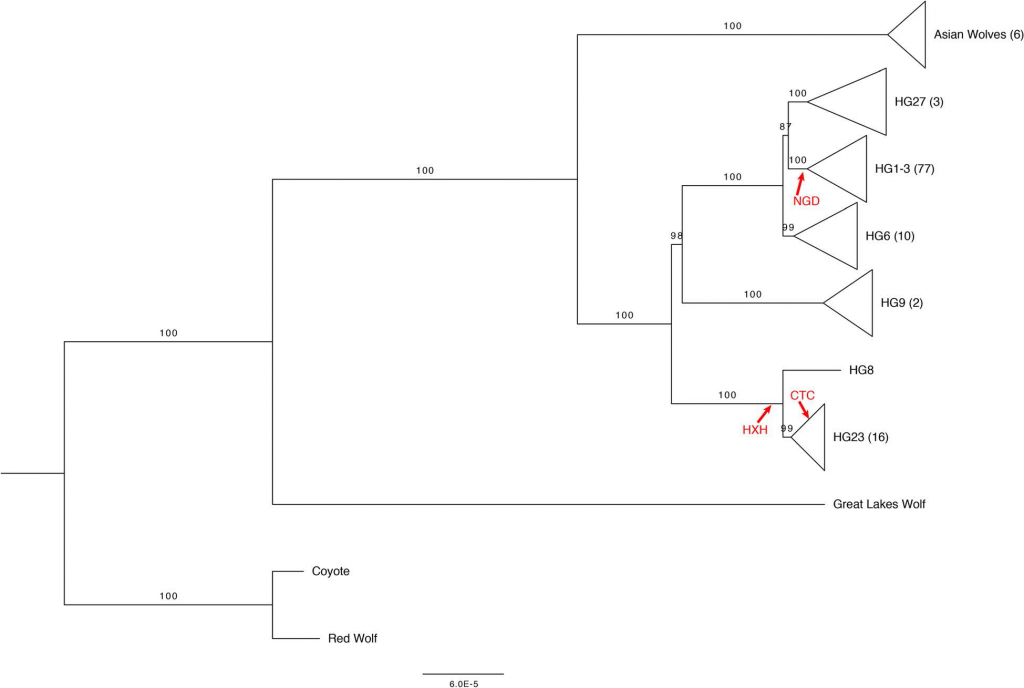
Using the variation discovered from sequence data, we applied a Bayesian MCMC approach to estimate TMRCAs for each haplotype group. Our estimated Y-chromosome mutation rate (3.07 × 10− 10 substitutions per site per year, relaxed clock model) falls within the range of a previous estimate by Ding et al. who used a similar calibration and estimate 1.35 × 10− 10– 4.31 × 10− 10 substitutions per site per year. The TMRCAs we estimated are substantially older than mitochondria phylogenies calibrated with tip dates of ancient samples, which report clade-specific TMRCAs < 25,000 years ago. We note that our Y-chromosome TMRCA estimates are extremely sensitive to our assumptions about the age of the root of the tree and should be interpreted with caution due to the uncertainty in this single calibration point. However, the relative ages of the branches and the chronological order of haplogroup divergences are more robust than the absolute estimated dates.
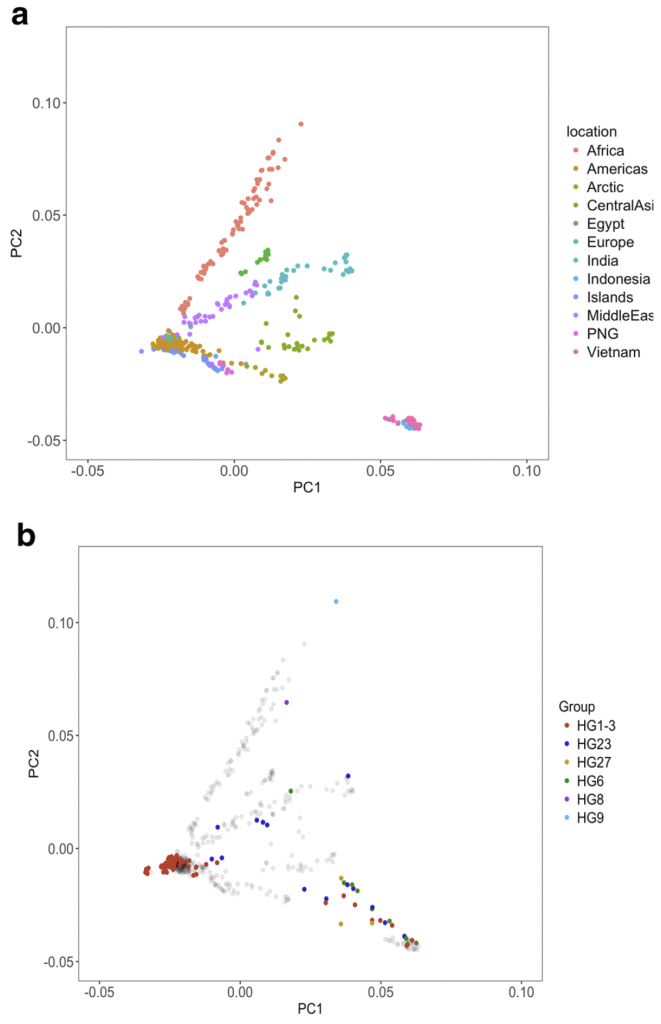
In general, the relationships between Y-chromosome haplogroups and autosomal ancestry we report are very similar to the relationships described in Shannon et al. As noted earlier, our dataset includes a subset of wolves with Y-chromosomes assigned to a dog Y-haplogroup. However, ADMIXTURE analysis does not indicate substantial recent dog ancestry in these samples, suggesting that their placement on the Y-chromosome phylogeny reflects variation in Y-chromosome haplotypes that was present in the ancestral population and therefore predates the domestication process or is the result of ancient introgression events whose signature of autosomal ancestry has been diluted.
Conclusions
Using sequencing data, we find that the estimated TMRCA of dog Y haplogroups predates dog domestication. We further reveal the placement of several wolf Y-chromosomes within deep branches of dog haplogroup clades. Using an expanded set of mutations diagnostic for each haplogroup, we find that distinct Y haplogroups were present in Europe during the Neolithic and that CTC, a ~ 4700 year old ancient dog from Germany has a Y-chromosome that shares diagnostic alleles with wolves found in India.
Other studies
On the same subject, you can read another recent study, bioRxiv preprint New Evidence of the Earliest Domestic Dogs in the Americas, by Perri et al. (2018); and also a recent, open access paper (see above featured image) Ancient European dog genomes reveal continuity since the Early Neolithic, by Botigué et al., Science Communications (2017).
While Proto-Indo-European- and Proto-Uralic-speakers had a close relationship with dogs (revealed in their reconstructed language and attributed archaeological cultures), I think it will be very difficult to ascertain any population movement based on them, unless there is a clear, well-established archaeological relationship between a specific culture and dog-breeding.
Nevertheless, I would say that this kind of studies are more likely to give some information related to these and other cultures than, for example, the study of honeybees in honey-hunting vs. beekeeping cultures (see e.g. The Complex Demographic History and Evolutionary Origin of the Western Honey Bee, Apis Mellifera, by Cridland, Tsutsui, and Ramírez GBE 2017), which was also related to the development of both PIE and PU cultures.
See also:
- Domesticated horse population structure, selection, and mtDNA geographic patterns
- Ancient DNA upends the horse family tree
- Consequences of Damgaard et al. 2018 (I): EHG ancestry in Maykop samples, and the potential Anatolian expansion routes
- No large-scale steppe migration into Anatolia; early Yamna migrations and MLBA brought LPIE dialects in Asia
- Domestication spread probably via the North Pontic steppe to Khvalynsk… but not horse riding
- Differing modes of animal exploitation in North Pontic Eneolithic and Bronze Age Societies
- Eneolithic Ukraine cultures of the North Pontic steppe and southern steppe-forest, on the Left Bank of the Dnieper
- Recent archaeological finds near Indo-European and Uralic homelands
- On the potential origin of Caucasus hunter-gatherer ancestry in Eneolithic steppe cultures