New preprint Population history from the Neolithic to present on the Mediterranean island of Sardinia: An ancient DNA perspective, by Marcus et al. bioRxiv (2019)
Interesting excerpts (emphasis mine, edited for clarity):
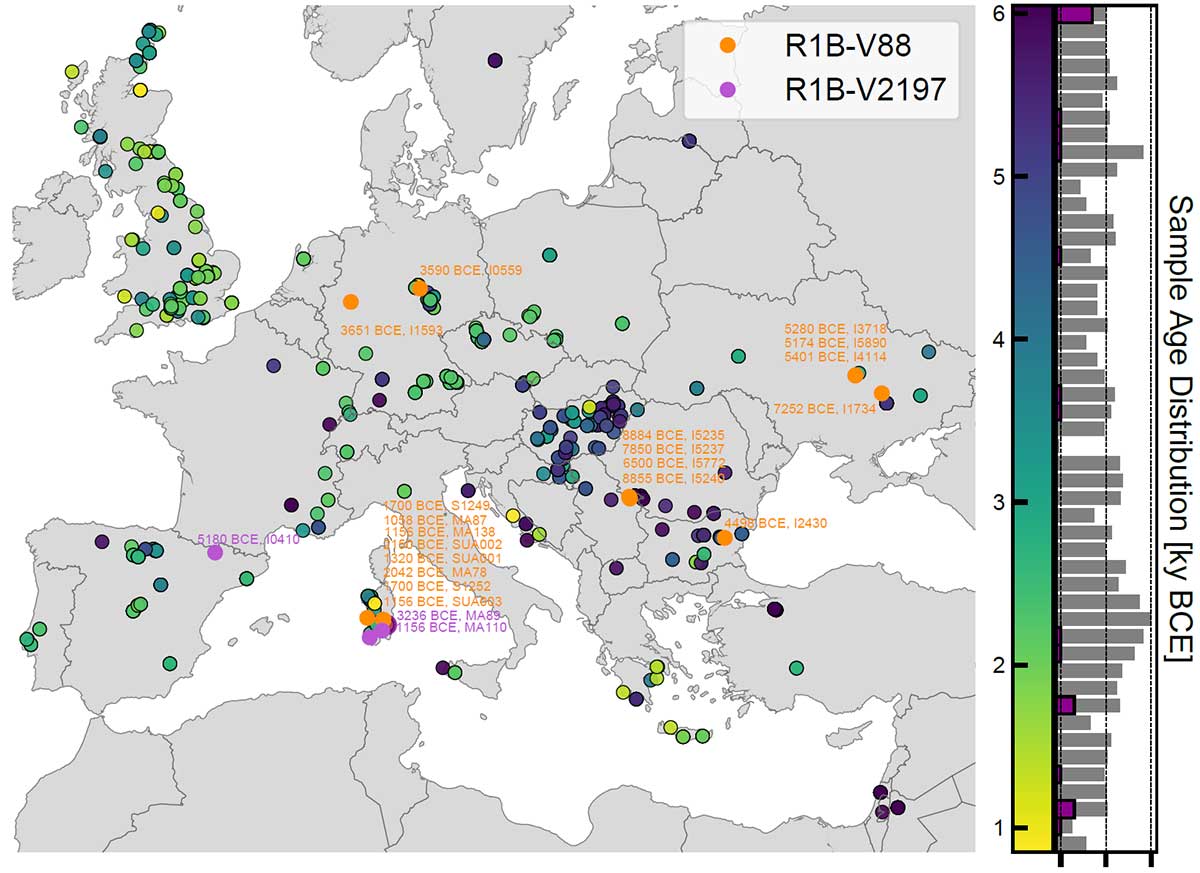
On the high frequency of R1b-V88
Our genome-wide data allowed us to assign Y haplogroups for 25 ancient Sardinian individuals. More than half of them consist of R1b-V88 (n=10) or I2-M223 (n=7).
Francalacci et al. (2013) identified three major Sardinia-specific founder clades based on present-day variation within the haplogroups I2-M26, G2-L91 and R1b-V88, and here we found each of those broader haplogroups in at least one ancient Sardinian individual. Two major present-day Sardinian haplogroups, R1b-M269 and E-M215, are absent.
Compared to other Neolithic and present-day European populations, the number of identified R1b-V88 carriers is relatively high.
(…)ancient Sardinian mtDNA haplotypes belong almost exclusively to macro-haplogroups HV (n = 16), JT (n = 17) and U (n = 9), a composition broadly similar to other European Neolithic populations.
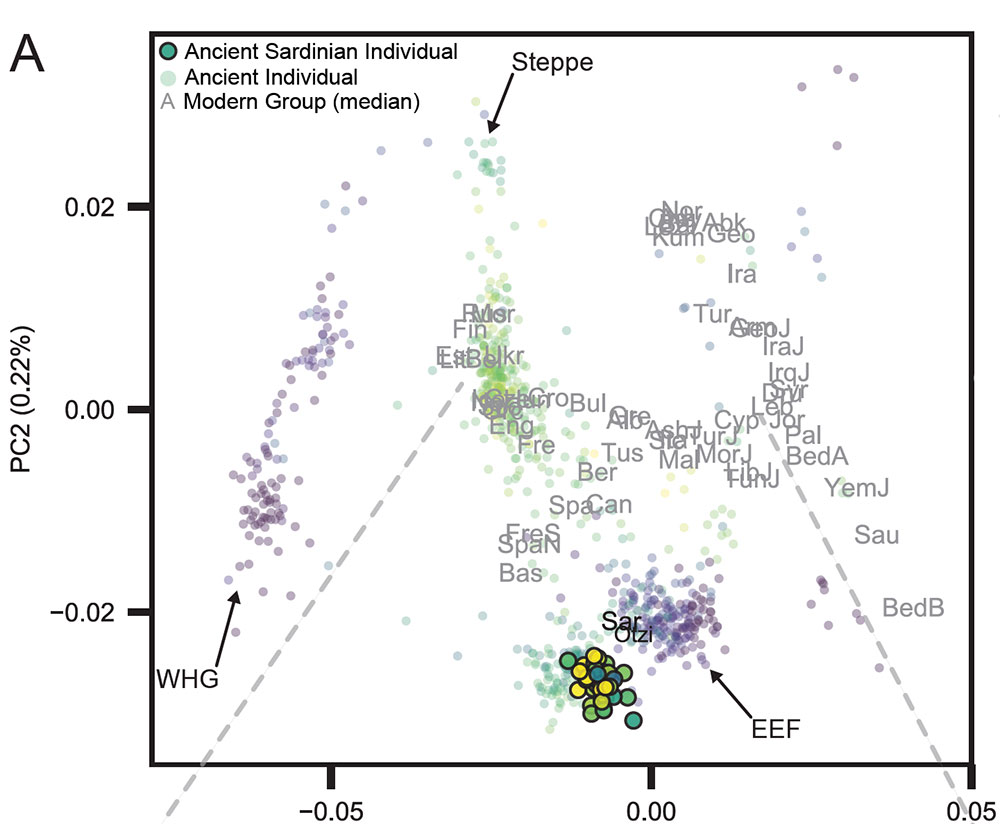
On the origin of a Vasconic-like Paleosardo with the Western EEF
(…) the Neolithic (and also later) ancient Sardinian individuals sit between early Neolithic Iberian and later Copper Age Iberian populations, roughly on an axis that differentiates WHG and EEF populations and embedded in a cluster that additionally includes Neolithic British individuals. This result is also evident in terms of absolute genetic differentiation, with low pairwise FST ~ 0.005 +- 0.002 between Neolithic Sardinian individuals and Neolithic western mainland European populations. Pairwise outgroup-f3 analysis shows a very similar pattern, with the highest values of f3 (i.e. most shared drift) being with Neolithic and Copper Age Iberia, gradually dropping off for temporally and geographically distant populations.
In explicit admixture models (using qpAdm, see Methods) the southern French Neolithic individuals (France-N) are the most consistent with being a single source for Neolithic Sardinia (p ~ 0:074 to reject the model of one population being the direct source of the other); followed by other populations associated with the western Mediterranean Neolithic Cardial Ware expansion.
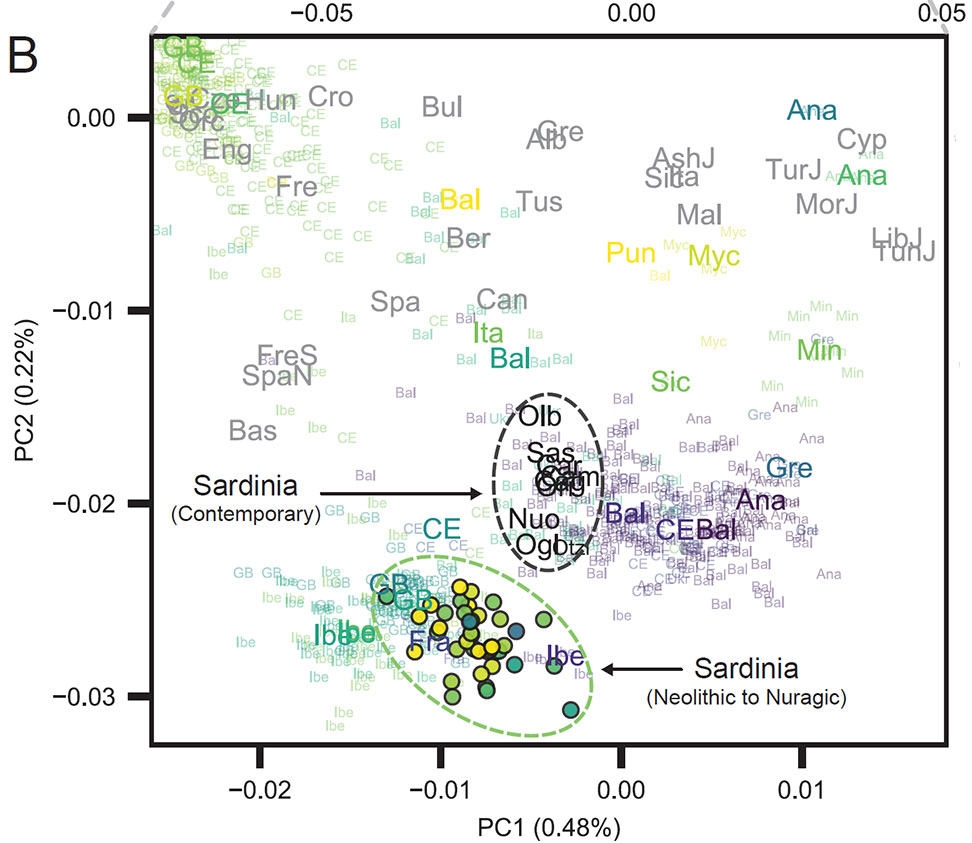
Pervasive Western Hunter-Gatherer ancestry in Iberian/French/Sardinian population
Similar to western European Neolithic and central European Late Neolithic populations, ancient Sardinian individuals are shifted towards WHG individuals in the top two PCs relative to early Neolithic Anatolians Admixture analysis using qpAdm infers that ancient Sardinian individuals harbour HG ancestry (~ 17%) that is higher than early Neolithic mainland populations (including Iberia, ~ 8%), but lower than Copper Age Iberians (~ 25%) and about the same as Southern French Middle-Neolithic individuals (~ 21%).
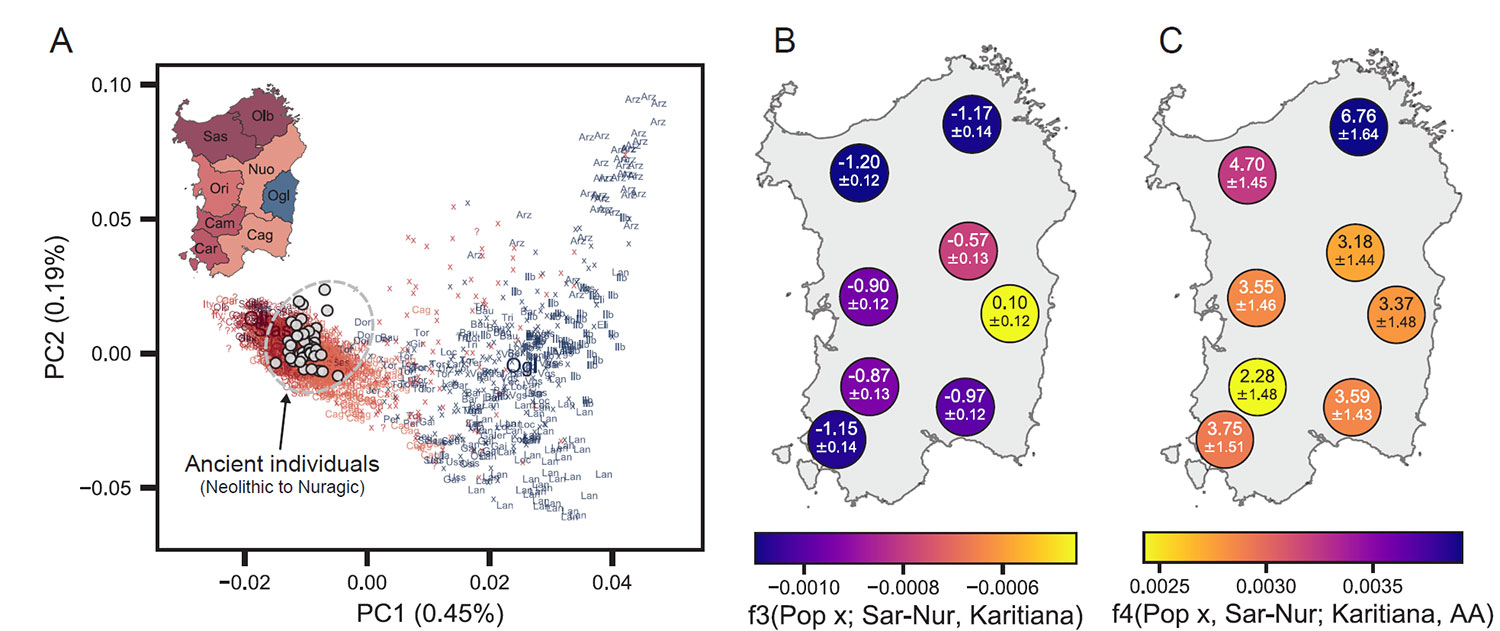
Continuity from Sardinia Neolithic through the Nuragic
We found several lines of evidence supporting genetic continuity from the Sardinian Neolithic into the Bronze Age and Nuragic times. Importantly, we observed low genetic differentiation between ancient Sardinian individuals from various time periods.
A qpAdm analysis, which is based on simultaneously testing f-statistics with a number of outgroups and adjusts for correlations, cannot reject a model of Neolithic Sardinian individuals being a direct predecessor of Nuragic Sardinian individuals (…) Our qpAdm analysis further shows that the WHG ancestry proportion, in a model of admixture with Neolithic Anatolia, remains stable at ~17% throughout three ancient time-periods.
Steppe influx in Modern Sardinians
While contemporary Sardinian individuals show the highest affinity towards EEF-associated populations among all of the modern populations, they also display membership with other clusters (Fig. 5). In contrast to ancient Sardinian individuals, present-day Sardinian individuals carry a modest “Steppe-like” ancestry component (but generally less than continental present-day European populations), and an appreciable broadly “eastern Mediterranean” ancestry component (also inferred at a high fraction in other present-day Mediterranean populations, such as Sicily and Greece).
Related
- Arrival of steppe ancestry with R1b-P312 in the Mediterranean: Balearic Islands, Sicily, and Iron Age Sardinia
- Y-DNA relevant in the postgenomic era, mtDNA study of Iron Age Italic population, and reconstructing the genetic history of Italians
- Modern Sardinians show elevated Neolithic farmer ancestry shared with Basques
- Ancient Phoenician mtDNA from Sardinia, Lebanon reflects settlement, genetic diversity, and female mobility
- Haplogroup J spread in the Mediterranean due to Phoenician and Greek colonizations
- Migration vs. Acculturation models for Aegean Neolithic in Genetics — still depending strongly on Archaeology
- Haplogroup R1b-M167/SRY2627 linked to Celts expanding with the Urnfield culture
- Aquitanians and Iberians of haplogroup R1b are exactly like Indo-Iranians and Balto-Slavs of haplogroup R1a
- Iberia: East Bell Beakers spread Indo-European languages; Celts expanded later
- Analysis of R1b-DF27 haplogroups in modern populations adds new information that contrasts with ‘steppe admixture’ results
- On Latin, Turkic, and Celtic – likely stories of mixed societies and little genetic impact
- First Iberian R1b-DF27 sample, probably from incoming East Bell Beakers
- Patterns of genetic differentiation and the footprints of historical migrations in the Iberian Peninsula
- First Iberian R1b-DF27 sample, probably from incoming East Bell Beakers
- Iberian prehistoric migrations in Genomics from Neolithic, Chalcolithic, and Bronze Age