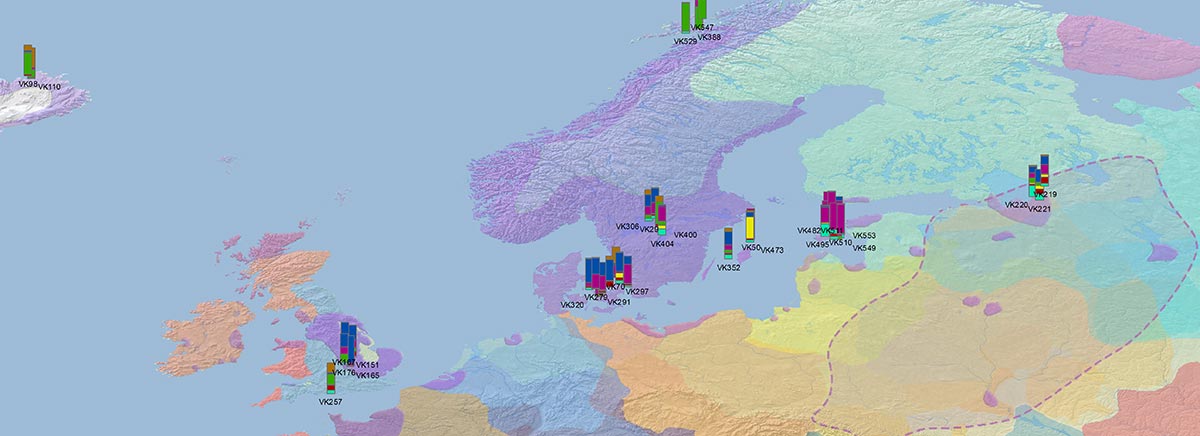
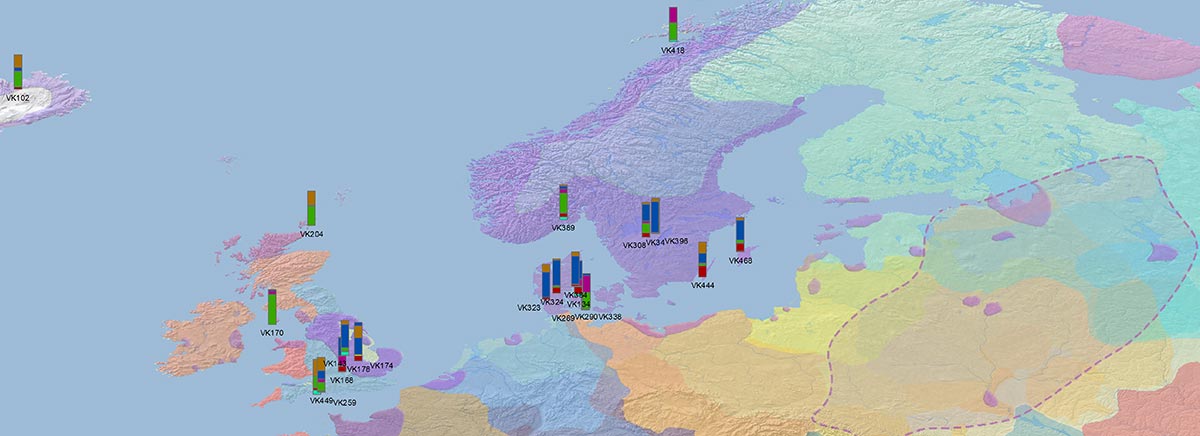
Recent paper (behind paywall) Population genomics of the Viking world, by Margaryan et al. Nature (2020), containing almost exactly the same information as its bioRxiv preprint.
I have used Y-SNP inferences recently reported by FTDNA (see below) to update my Ancient DNA Dataset and the ArcGIS Online Map, and also to examine the chronological and geographical evolution of Y-DNA (alone and in combination with ancestry).
Sections of this post:
- Iron Age to Medieval Y-DNA
- Iron Age to Medieval Ancestry
- Iron Age to Medieval Y-DNA + Ancestry
- FTDNA’s big public debut
I. Iron Age to Medieval Y-DNA
#Vikings ancient DNA mapped through the evolution of Y-chromosome haplogroups, from data in Margaryan et al. (Nature) 2020 supp. tables with many Y-SNP calls by FTDNA, and other reported ancient samples…(2/3) pic.twitter.com/kjaf05I5qj
— Carlos Quiles (@cquilesc) September 24, 2020
These are some selected periods in PDF (to better appreciate the evolution of individual samples), in 100 year intervals:
 Ancient Y-DNA ca. AD 250-350 (PDF)
Ancient Y-DNA ca. AD 250-350 (PDF) Ancient Y-DNA ca. AD 350-450 (PDF)
Ancient Y-DNA ca. AD 350-450 (PDF) Ancient Y-DNA ca. AD 450-550 (PDF)
Ancient Y-DNA ca. AD 450-550 (PDF) Ancient Y-DNA ca. AD 550-650 (PDF)
Ancient Y-DNA ca. AD 550-650 (PDF) Ancient Y-DNA ca. AD 650-750 (PDF)
Ancient Y-DNA ca. AD 650-750 (PDF) Ancient Y-DNA ca. AD 750-850 (PDF)
Ancient Y-DNA ca. AD 750-850 (PDF) Ancient Y-DNA ca. AD 850-950 (PDF)
Ancient Y-DNA ca. AD 850-950 (PDF) Ancient Y-DNA ca. AD 950-1050 (PDF)
Ancient Y-DNA ca. AD 950-1050 (PDF) Ancient Y-DNA ca. AD 1050-1150 (PDF)
Ancient Y-DNA ca. AD 1050-1150 (PDF) Ancient Y-DNA ca. AD 1150-1250 (PDF)
Ancient Y-DNA ca. AD 1150-1250 (PDF) Ancient Y-DNA ca. AD 1250-1350 (PDF)
Ancient Y-DNA ca. AD 1250-1350 (PDF) Ancient ancestry ca. AD 1350-1450 (PDF)
Ancient ancestry ca. AD 1350-1450 (PDF)
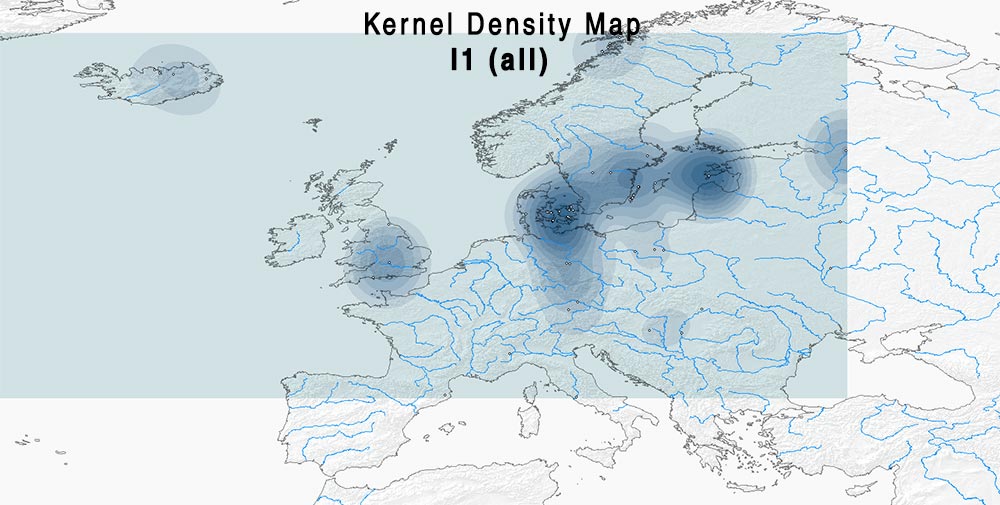
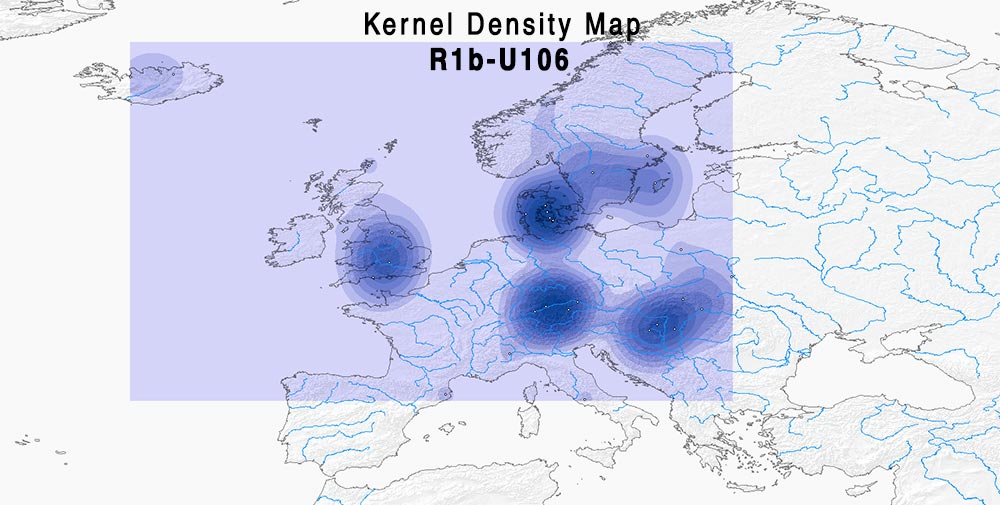
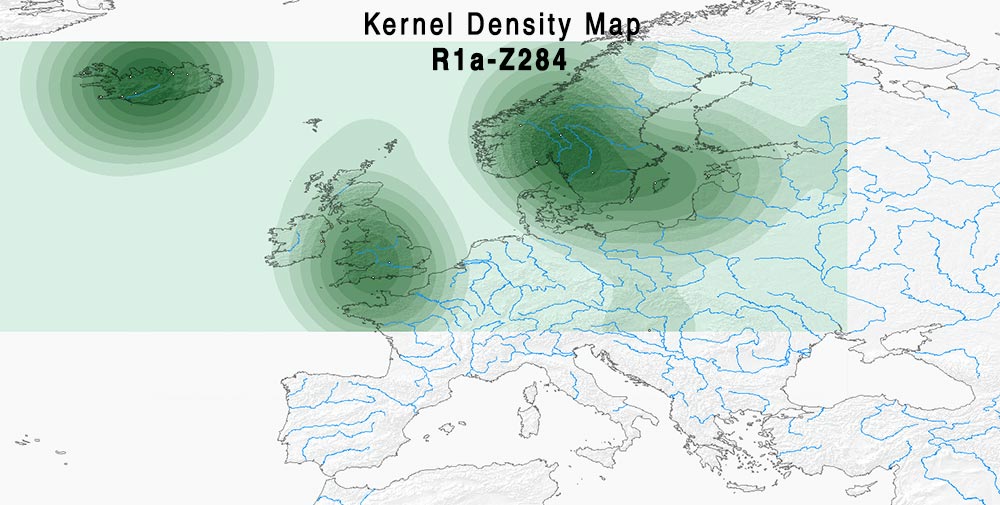
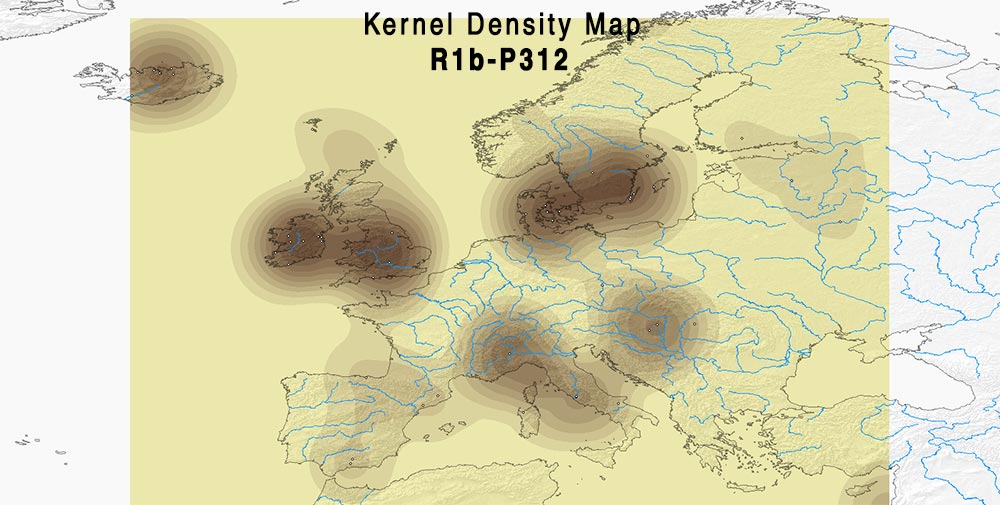
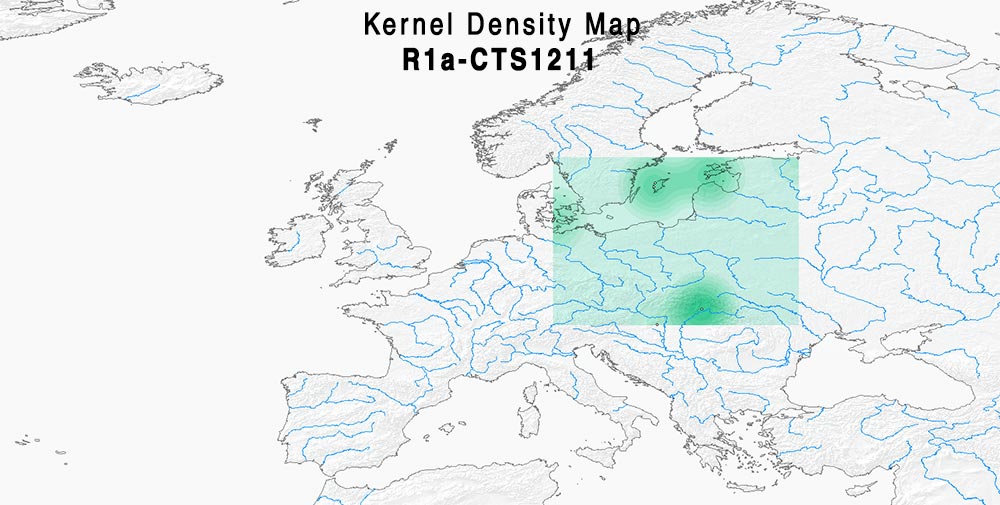
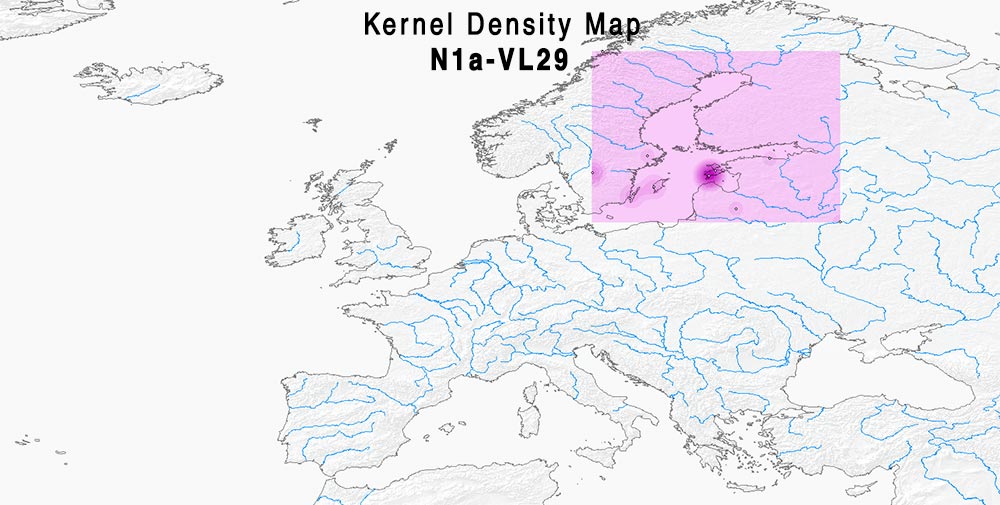
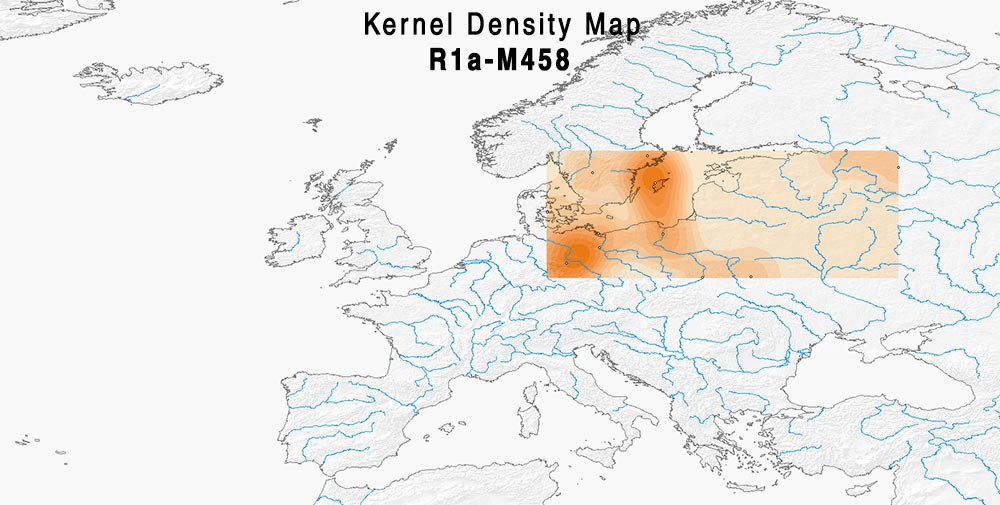
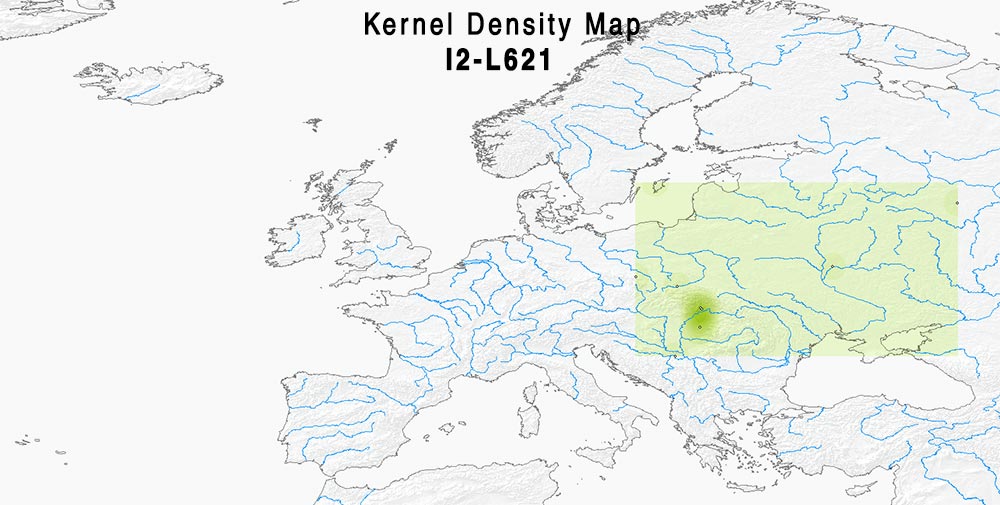
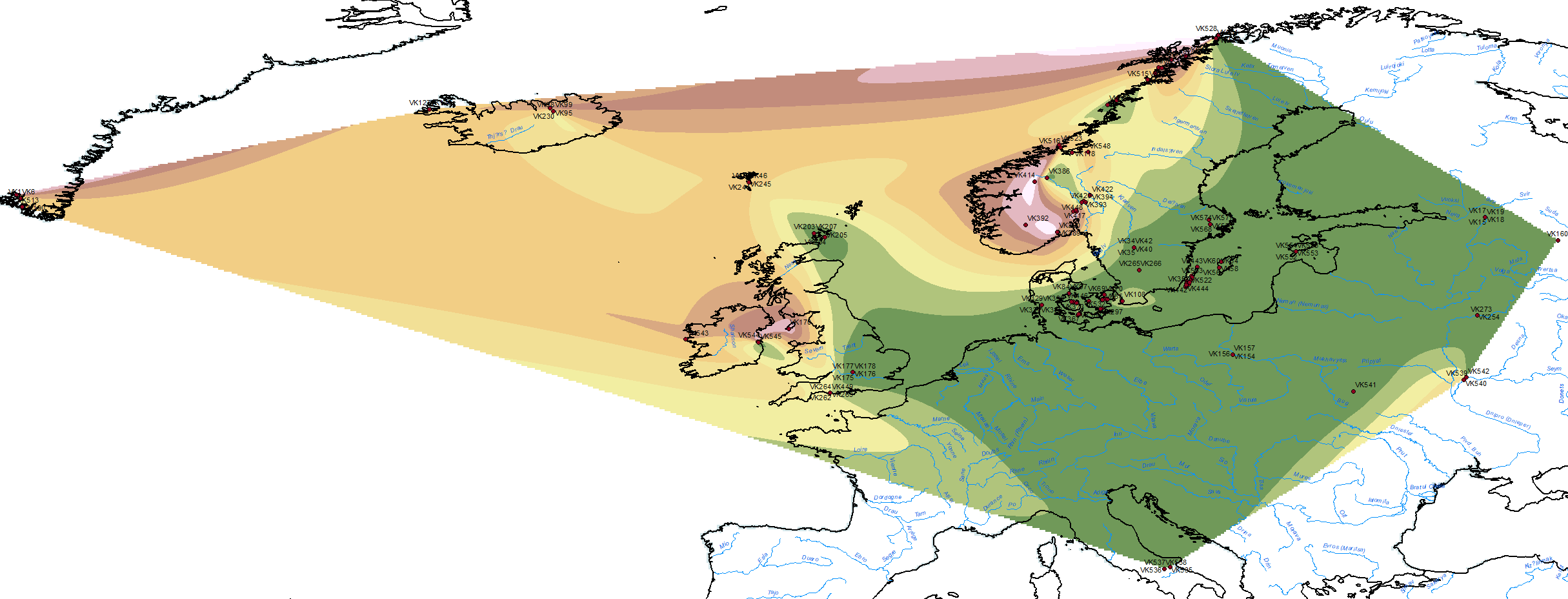
Here are some relevant Kernel Density maps obtained by selecting specific Y-DNA haplogroups from the Late Iron Age to the Middle Ages (ca. AD 400-1300). The main problem with these “heat maps” is the lack of proper random geographical sampling.
II. Iron Age to Medieval Ancestry
A word of caution is due (same as last year): by selecting modern populations – some of them closely related – to test Iron Age and Medieval samples, the authors have consciously chosen to draw an anachronistic picture prone to misinterpretations, even though it might also be useful to generate new hypotheses to be tested.
#Vikings ancient DNA mapped through the ancestry of modern populations, from data in Margaryan et al. (Nature) 2020 supp. tables https://t.co/Ow5BgTyBHc…(1/3) pic.twitter.com/5Xy1ZhTodh
— Carlos Quiles (@cquilesc) September 24, 2020
This is how I would interpret each ancestry at first sight, assuming that some or all of these components must have formed well after the first Viking expansions:
- “Modern British”: Based on the Iron Age “Pict” from Orkney (ca. AD 500), this is probably a good proxy for pre-Germanic populations of Great Britain and Ireland, although it also likely includes ancestry proper of ancient populations from North-West Europe.
- “Modern Swedish”: Based on early samples from Denmark, from Sweden (VK522) and Norway (VK418), it is a potential good proxy for East Scandinavian ancestry, but it seems to be present also in ‘local’ samples from the North-East Baltic.
- “Modern Norwegian”: Based on early samples from Norway, this also looks like a decent proxy for ancient West Scandinavian populations.
- “Modern Danish”: Based on the Iron Age sample from Zealand (VK521) and a later one from Funen (VK301) showing varied admixture, as well as later ones from Southern Scandinavia showing a more and more homogeneous “Danish” ancestry, it looks like it is not a reliable proxy for Iron Age peoples from Jutland.
- “Modern Polish”: There are no early samples with this ancestry in the paper, and the earliest to appear show other components, so its relationship to early Slavic ancestry is unclear. It is found on both ends of the Baltic-related West to East Slavic spectrum during the Middle Ages, so it certainly looks like a good proxy for modern northern Slavs.
- “Modern Italian”: since it appears everywhere among northern European samples, it is probably also a proxy for Central European peoples, not only Southern ones. Hence, ancient samples of West Germanic origin would probably show different mixtures of “Modern Danish”, “Modern British”, and “Modern Italian”.
- “Modern Finnish”: probably a reasonable proxy for medieval (South-West?)-Finnish-like ancestry, although its association with “Modern Swedish” and “Modern Polish” makes all three not very informative as to the potential links with ancient Balto-Slavic- or Germanic-speaking peoples vs. Finnic-speaking ones.
These are some selected periods in PDF (to better appreciate the evolution of individual samples), in 100 year intervals:
 Ancient ancestry ca. AD 250-350 (PDF)
Ancient ancestry ca. AD 250-350 (PDF) Ancient ancestry ca. AD 350-450 (PDF)
Ancient ancestry ca. AD 350-450 (PDF) Ancient ancestry ca. AD 450-550 (PDF)
Ancient ancestry ca. AD 450-550 (PDF) Ancient ancestry ca. AD 550-650 (PDF)
Ancient ancestry ca. AD 550-650 (PDF) Ancient ancestry ca. AD 650-750 (PDF)
Ancient ancestry ca. AD 650-750 (PDF) Ancient ancestry ca. AD 750-850 (PDF)
Ancient ancestry ca. AD 750-850 (PDF) Ancient ancestry ca. AD 850-950 (PDF)
Ancient ancestry ca. AD 850-950 (PDF) Ancient ancestry ca. AD 950-1050 (PDF)
Ancient ancestry ca. AD 950-1050 (PDF) Ancient ancestry ca. AD 1050-1150 (PDF)
Ancient ancestry ca. AD 1050-1150 (PDF) Ancient ancestry ca. AD 1150-1250 (PDF)
Ancient ancestry ca. AD 1150-1250 (PDF) Ancient ancestry ca. AD 1250-1350 (PDF)
Ancient ancestry ca. AD 1250-1350 (PDF) Ancient ancestry ca. AD 1350-1450 (PDF)
Ancient ancestry ca. AD 1350-1450 (PDF) Ancient ancestry general (PDF)
Ancient ancestry general (PDF)
For maps of neighbour interpolation of the reported ancestry, see my previous post Vikings, Vikings, Vikings! “eastern” ancestry in the whole Baltic Iron Age.
III. Iron Age to Medieval Ancestry + Y-DNA
This is probably the trickiest part. There is no obvious relationship between Y-DNA and ancestry. Nevertheless, whenever we are lucky enough to get a proper sampling of a certain period, we might occasionally find some relevant links between the expansion of very specific subclades and a certain admixture.
Here are some maps that might be useful to assess that:
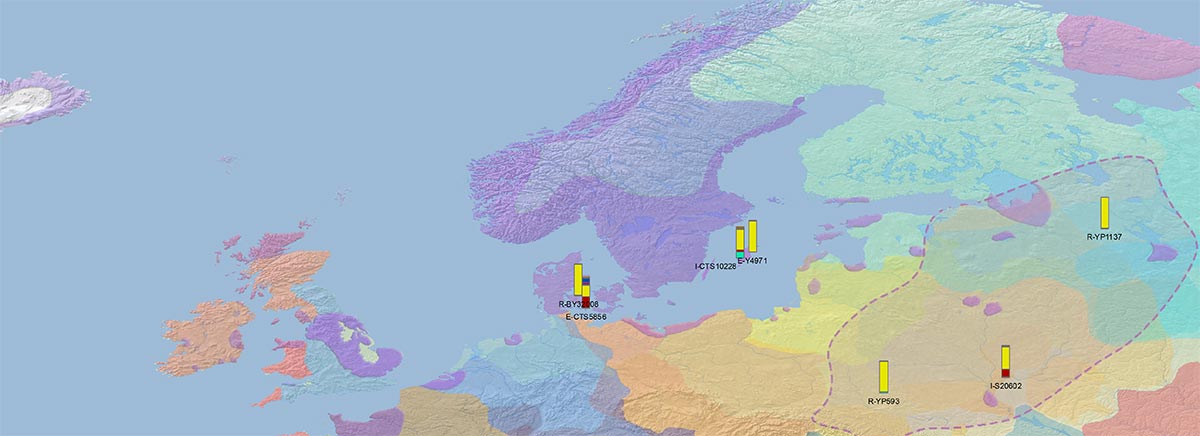
NOTE. I usually labelled samples with their ID, but occasionally the terminal SNPs are selected for clarity.
Admixture and haplogroups I1, I2
The division of I1 into ancient branches is probably too simplistic, and would need a careful selection of recent ones to see if regional patterns emerge:
 Admixture of samples of hg. I1a1 (PDF)
Admixture of samples of hg. I1a1 (PDF) Admixture of samples of hg. I1a2 (PDF)
Admixture of samples of hg. I1a2 (PDF) Admixture of samples of hg. I1 (xI1a1, xI1a2) (PDF)
Admixture of samples of hg. I1 (xI1a1, xI1a2) (PDF) Admixture of samples of hg. I2 (xL621) (PDF)
Admixture of samples of hg. I2 (xL621) (PDF)
Admixture and R1b subclades
If modern ancestry components can be trusted, it looks like R1b-U106 and R1b-U152 also need a careful selection of subclades to see if there is some relationship between regions and ancestral components, whereas R1b-Z290 (including the common R-L21 subclade) is more clearly associated (at least the earlier samples) with “British”-like admixture.
 Admixture of samples of hg. R1b-U106 (PDF)
Admixture of samples of hg. R1b-U106 (PDF) Admixture of samples of hg. R1b-Z290 (PDF)
Admixture of samples of hg. R1b-Z290 (PDF) Admixture of samples of hg. R1b-U152 (PDF)
Admixture of samples of hg. R1b-U152 (PDF) Admixture of samples of hg. R1b-DF27 (PDF)
Admixture of samples of hg. R1b-DF27 (PDF) Admixture of samples of hg. R1b-P312(xZ290, xZZ11) or L151(xP312) (PDF).
Admixture of samples of hg. R1b-P312(xZ290, xZZ11) or L151(xP312) (PDF).
Admixture and R1a and N1a subclades
The case of R1a-Z284 looks probably more “Norwegian” and “Swedish”-shifted – i.e. Northern Scandinavian – than I1 and R1b-U106 above, whereas R1a-Z280 looks more East Baltic-related.
In fact, a curious finding by FTDNA is the Y-DNA connection of newly reported R1a-CTS1211 ‘basal’ subclade R-YP4932, shared by VK487 from Saaremaa (ca. AD 750) with Estonian LBA samples V9_2 from Joelähtme (ca. 1115 BC) and X14_1 from Rebala (ca. 605 BC), both from the Harju region, which attests to the continuity of male lineages (and not only ancestry) among Baltic Finns around the Gulf of Finland.
#EDIT: On the other hand, depending on how one interprets those “Swedish”-like Vikings of hg. N-VL29 from Estonia and Northern Russia, they could also be interpreted as a back-migration of lineages associated with Germanic-speaking Scandinavians since the Early Iron Age…
 Admixture of samples of hg. R1a-Z284 (PDF)
Admixture of samples of hg. R1a-Z284 (PDF) Admixture of samples of hg. R1a-M458, I2-L621, E1b (PDF)
Admixture of samples of hg. R1a-M458, I2-L621, E1b (PDF) Admixture of samples of hg. R1a-CTS1211 and R1a-Z92 (PDF)
Admixture of samples of hg. R1a-CTS1211 and R1a-Z92 (PDF) Admixture of samples of hg. N1a-VL29 (most N1a-L550) (PDF)
Admixture of samples of hg. N1a-VL29 (most N1a-L550) (PDF) Admixture of samples of varied haplogroups (G2a, J2a, J2b, Z2103, T1a). (PDF)
Admixture of samples of varied haplogroups (G2a, J2a, J2b, Z2103, T1a). (PDF)
I have put together the most commonly assumed Early Slavic Y-DNA markers (that is, I2-L621, E1b-V13, and R1a-M458) in one of the maps, to show their recent connection with “Modern Polish” ancestry.
IV. FTDNA’s big public debut
FamilyTreeDNA has published in a matter of days detailed Y-SNP inferences and useful comments for 285 samples from those released at ENA, as reported step by step in Roberta Estes’ blog DNAeXplained. If you are an FTDNA client and lucky enough to have a distant family member among those sampled, you will find them in your tree by now.
The FTDNA team formed by Michael Sager and Göran Runfeldt – the same pair that contributed to the recent paper on the Hungarian Árpád Dynasty – had been already testing all ancient samples available for many weeks now, adding many of them to the FTDNA Haplotree.
NOTE. Up until now, those additions had been more or less hidden to the general public, except for some cases that have been announced by Estes or Runfeldt – and I have included in the Ancient DNA Dataset. Some of those still hidden are known to be there from their likely position and the new branches formed. As far as I could see, there are some other more or less obvious ancient samples in the haplotree, which I haven’t included because I can’t be sure.
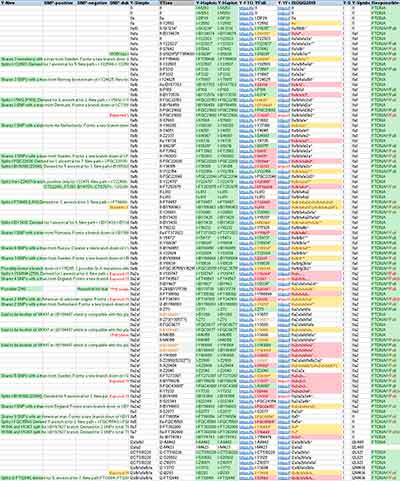
As you might notice from the Ancient DNA Dataset, it is slowly but radically changing the visual output into a sea of Y-SNPs with green background that reflects FTDNA’s seal of approval. Advantages over what we had before include:
1) The combination of coverage and quality accepted by FTDNA has a lower threshold than the one used by YFull, which means that we get confirmation of SNPs from lower coverage samples which, even though not very informative from a genealogical perspective, can be very interesting from a (pre)historical point of view.
2) The pace at which FTDNA can manually check Y-SNPs clearly beats any other lab or amateur. As of yesterday, when I finished getting samples from both, YFull had analysed half or less, had some of them still in progress, and there are probably still some with good enough coverage to be included.
3) The depth of assignment in FTDNA is usually one step or more beyond YFull’s thanks to their larger database (by 50% or more) – in my dataset, one subclade behind by YFull is in yellow background, two subclades or more in magenta background. Many samples have split branches only present in FTDNA, which cannot be followed by YFull.
NOTE. FTDNA is said to be slightly more “Eurocentric” – which means that its database is even larger in terms of common European haplogroups – but so is (and will probably remain) the ancient DNA record.
4) The quality of SNP inferences is also astonishing. A simple glance at the dataset (see only Vikings) will show you some weird misplacements by YFull (which I marked in red). This is probably due to (a) the lack of a proper SNP database in some cases (e.g. for I1 or below ZZ11, a pattern also seen for years in comparison with YTree), (b) the competition that both teams have probably had to include these samples in their trees as quickly as possible, and (c) the unmatched experience of the FTDNA team.
NOTE. In two or three cases, YFull has selected one or more SNPs further down the one reported by FTDNA, which seems to correspond to controversial – although possible – calls (marked like other estimations). In similar cases, FTDNA informs of that potential downstream subclade in their comments.
I hope YFull will eventually try to compete in quality and quantity, so that we all benefit in the end, but in the meantime Family Tree DNA is clearly the new standard for assessing ancient DNA.
That new standard also includes tracking ancient SNP migrations and TMRCA estimates with Rob Spencer’s SNP tracker, and tree relationships with his SNP Tree Explorer, the only available free tools that offer a professional level assessment of Y-DNA migration paths and chronology (with Alex Williamson’s YTree becoming outdated as of late).
NOTE. I am in no way affiliated with FTDNA, nor do I get any money or benefit from this hobby at all. Rob Spencer also doesn’t benefit in any way from his work. I am not interested in the usual “genetic lab wars” that people so often join; just in seeing better SNP inferences and, if possible, a nice and healthy competition. If you have read this blog for some time now, you know I have usually linked to YFull Y-SNP calls and estimates, and I will keep doing so.
See also
- Vikings, Vikings, Vikings! “eastern” ancestry in the whole Baltic Iron Age
- “Local” Tollense Valley warriors linked to Germanic peoples
- Tug of war between Balto-Slavic and West Uralic (II)
- European hydrotoponymy (III): from Old European to Palaeo-Germanic and the Nordwestblock
- Genetic continuity among Uralic-speaking cultures in north-eastern Europe
- European hydrotoponymy (IV): tug of war between Balto-Slavic and West Uralic
- Pre-Germanic and Pre-Balto-Finnic shared vocabulary from Pitted Ware seal hunters
- Pre-Germanic born out of a Proto-Finnic substrate in Scandinavia
- Bell Beaker/early Late Neolithic (NOT Corded Ware/Battle Axe) identified as forming the Pre-Germanic community in Scandinavia
- Reproductive success among ancient Icelanders stratified by ancestry
- Genomic analysis of Germanic tribes from Bavaria show North-Central European ancestry
- Germanic tribes during the Barbarian migrations show mainly R1b, also I lineages
- Minimal Corded Ware culture impact in Scandinavia – Bell Beakers the unifying maritime elite